The Altair Community is migrating to a new platform to provide a better experience for you. In preparation for the migration, the Altair Community is on read-only mode from October 28 - November 6, 2024. Technical support via cases will continue to work as is. For any urgent requests from Students/Faculty members, please submit the form linked here
How to Plot multi-class ROC curve in Rapidminer?
 Learner III
Learner III
How to plot the Multiclass ROC curve from below details (Results) in one Graph! There are Six Classes in my Case. A Picture is also uploaded for Better understanding of Multiclass ROC curves in One graph for an algorithm.

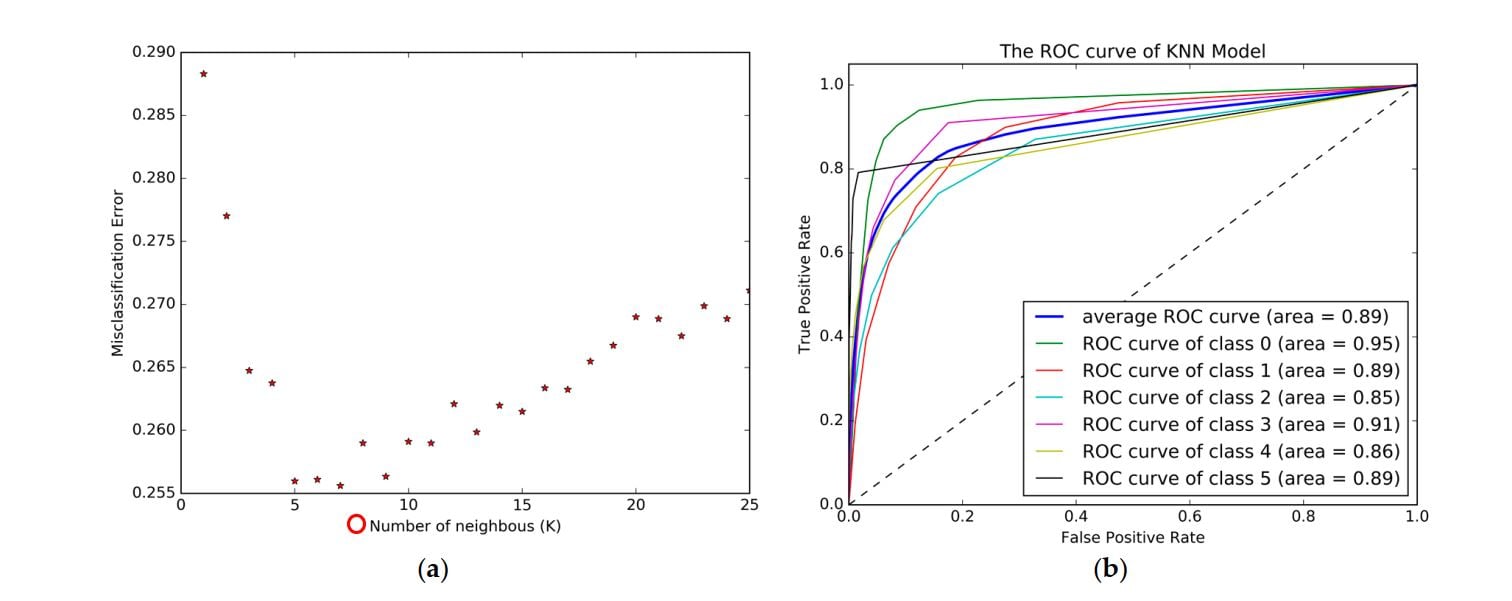
Regards


Tagged:
0
Best Answers
-
 Telcontar120
RapidMiner Certified Analyst, RapidMiner Certified Expert, Member Posts: 1,635
Telcontar120
RapidMiner Certified Analyst, RapidMiner Certified Expert, Member Posts: 1,635  Unicorn
There is really no such thing as a multiclass ROC curve. Each of of these curves is likely generated by a "one vs all other" approach (e.g., when looking at class A, the curve is a vs not-A, and so on). You can only do this manually in RapidMiner by linking multiple performance operators after building different models on different labels (you need a binominal label for each class using the one vs all others approach). Then you can have a curve for each of the classes separately).
Unicorn
There is really no such thing as a multiclass ROC curve. Each of of these curves is likely generated by a "one vs all other" approach (e.g., when looking at class A, the curve is a vs not-A, and so on). You can only do this manually in RapidMiner by linking multiple performance operators after building different models on different labels (you need a binominal label for each class using the one vs all others approach). Then you can have a curve for each of the classes separately).
@sgenzer I think Compare ROC doesn't help in this circumstances since it requires you to have the same label for all the inner learners which are run at the same time.
But if someone else knows a better way to do what the OP is asking, I would be very interested.6 -
 varunm1
Member Posts: 1,207
varunm1
Member Posts: 1,207  Unicorn
@MunchCrunch19
Unicorn
@MunchCrunch19
you need to manually relabel and generate multiple datasets,
For example, you have three classes A, B, C. In the first dataset you will keep A but B and C are relabelled as D (now it is converted to binomial (A and D ). Similarly, you will keep B and relabel A and C as E which gives you a second dataset with classes B and E and soon. As you have 6 classes you will have 6 datasets.
There might be other better and simple ways. But this is what I did using IRIS dataset that has 3 classes. Check the expression in generate attribute where I am relabelling and converting to binomial classification. XML code below. The figure for reference below. You get multiple AUC values.<?xml version="1.0" encoding="UTF-8"?><process version="9.2.001">
<context>
<input/>
<output/>
<macros/>
</context>
<operator activated="true" class="process" compatibility="9.2.001" expanded="true" name="Process">
<parameter key="logverbosity" value="init"/>
<parameter key="random_seed" value="2001"/>
<parameter key="send_mail" value="never"/>
<parameter key="notification_email" value=""/>
<parameter key="process_duration_for_mail" value="30"/>
<parameter key="encoding" value="SYSTEM"/>
<process expanded="true">
<operator activated="true" class="retrieve" compatibility="9.2.001" expanded="true" height="68" name="Retrieve Iris" width="90" x="45" y="85">
<parameter key="repository_entry" value="//Samples/data/Iris"/>
</operator>
<operator activated="true" class="multiply" compatibility="9.2.001" expanded="true" height="124" name="Multiply" width="90" x="179" y="85"/>
<operator activated="true" class="generate_attributes" compatibility="9.2.001" expanded="true" height="82" name="Generate Attributes (2)" width="90" x="313" y="136">
<list key="function_descriptions">
<parameter key="Binomial_Labels_Generated" value="if(label == "Iris-virginica", "A", (if(label == "Iris-versicolor","A",label)))"/>
</list>
<parameter key="keep_all" value="true"/>
</operator>
<operator activated="true" class="generate_attributes" compatibility="9.2.001" expanded="true" height="82" name="Generate Attributes" width="90" x="313" y="34">
<list key="function_descriptions">
<parameter key="Binomial_Labels_Generated" value="if(label == "Iris-virginica", "A", (if(label == "Iris-setosa","A",label)))"/>
</list>
<parameter key="keep_all" value="true"/>
</operator>
<operator activated="true" class="select_attributes" compatibility="9.2.001" expanded="true" height="82" name="Select Attributes" width="90" x="447" y="34">
<parameter key="attribute_filter_type" value="single"/>
<parameter key="attribute" value="label"/>
<parameter key="attributes" value=""/>
<parameter key="use_except_expression" value="false"/>
<parameter key="value_type" value="attribute_value"/>
<parameter key="use_value_type_exception" value="false"/>
<parameter key="except_value_type" value="time"/>
<parameter key="block_type" value="attribute_block"/>
<parameter key="use_block_type_exception" value="false"/>
<parameter key="except_block_type" value="value_matrix_row_start"/>
<parameter key="invert_selection" value="true"/>
<parameter key="include_special_attributes" value="true"/>
</operator>
<operator activated="true" class="set_role" compatibility="9.2.001" expanded="true" height="82" name="Set Role" width="90" x="581" y="34">
<parameter key="attribute_name" value="Binomial_Labels_Generated"/>
<parameter key="target_role" value="label"/>
<list key="set_additional_roles"/>
</operator>
<operator activated="true" class="concurrency:cross_validation" compatibility="9.2.001" expanded="true" height="145" name="Cross Validation" width="90" x="715" y="34">
<parameter key="split_on_batch_attribute" value="false"/>
<parameter key="leave_one_out" value="false"/>
<parameter key="number_of_folds" value="10"/>
<parameter key="sampling_type" value="automatic"/>
<parameter key="use_local_random_seed" value="false"/>
<parameter key="local_random_seed" value="1992"/>
<parameter key="enable_parallel_execution" value="true"/>
<process expanded="true">
<operator activated="true" class="concurrency:parallel_decision_tree" compatibility="9.2.001" expanded="true" height="103" name="Decision Tree" width="90" x="112" y="34">
<parameter key="criterion" value="gain_ratio"/>
<parameter key="maximal_depth" value="10"/>
<parameter key="apply_pruning" value="true"/>
<parameter key="confidence" value="0.1"/>
<parameter key="apply_prepruning" value="true"/>
<parameter key="minimal_gain" value="0.01"/>
<parameter key="minimal_leaf_size" value="2"/>
<parameter key="minimal_size_for_split" value="4"/>
<parameter key="number_of_prepruning_alternatives" value="3"/>
</operator>
<connect from_port="training set" to_op="Decision Tree" to_port="training set"/>
<connect from_op="Decision Tree" from_port="model" to_port="model"/>
<portSpacing port="source_training set" spacing="0"/>
<portSpacing port="sink_model" spacing="0"/>
<portSpacing port="sink_through 1" spacing="0"/>
</process>
<process expanded="true">
<operator activated="true" class="apply_model" compatibility="9.2.001" expanded="true" height="82" name="Apply Model" width="90" x="112" y="34">
<list key="application_parameters"/>
<parameter key="create_view" value="false"/>
</operator>
<operator activated="true" class="performance_binominal_classification" compatibility="9.2.001" expanded="true" height="82" name="Performance" width="90" x="246" y="34">
<parameter key="main_criterion" value="first"/>
<parameter key="accuracy" value="true"/>
<parameter key="classification_error" value="false"/>
<parameter key="kappa" value="false"/>
<parameter key="AUC (optimistic)" value="false"/>
<parameter key="AUC" value="true"/>
<parameter key="AUC (pessimistic)" value="false"/>
<parameter key="precision" value="false"/>
<parameter key="recall" value="false"/>
<parameter key="lift" value="false"/>
<parameter key="fallout" value="false"/>
<parameter key="f_measure" value="false"/>
<parameter key="false_positive" value="false"/>
<parameter key="false_negative" value="false"/>
<parameter key="true_positive" value="false"/>
<parameter key="true_negative" value="false"/>
<parameter key="sensitivity" value="false"/>
<parameter key="specificity" value="false"/>
<parameter key="youden" value="false"/>
<parameter key="positive_predictive_value" value="false"/>
<parameter key="negative_predictive_value" value="false"/>
<parameter key="psep" value="false"/>
<parameter key="skip_undefined_labels" value="true"/>
<parameter key="use_example_weights" value="true"/>
</operator>
<connect from_port="model" to_op="Apply Model" to_port="model"/>
<connect from_port="test set" to_op="Apply Model" to_port="unlabelled data"/>
<connect from_op="Apply Model" from_port="labelled data" to_op="Performance" to_port="labelled data"/>
<connect from_op="Performance" from_port="performance" to_port="performance 1"/>
<portSpacing port="source_model" spacing="0"/>
<portSpacing port="source_test set" spacing="0"/>
<portSpacing port="source_through 1" spacing="0"/>
<portSpacing port="sink_test set results" spacing="0"/>
<portSpacing port="sink_performance 1" spacing="0"/>
<portSpacing port="sink_performance 2" spacing="0"/>
</process>
</operator>
<operator activated="true" class="converters:roc_curve_2_example_set" compatibility="0.5.000" expanded="true" height="82" name="ROC Curve to ExampleSet" width="90" x="849" y="34">
<parameter key="Number of Points" value="500"/>
</operator>
<operator activated="true" class="select_attributes" compatibility="9.2.001" expanded="true" height="82" name="Select Attributes (2)" width="90" x="447" y="187">
<parameter key="attribute_filter_type" value="single"/>
<parameter key="attribute" value="label"/>
<parameter key="attributes" value=""/>
<parameter key="use_except_expression" value="false"/>
<parameter key="value_type" value="attribute_value"/>
<parameter key="use_value_type_exception" value="false"/>
<parameter key="except_value_type" value="time"/>
<parameter key="block_type" value="attribute_block"/>
<parameter key="use_block_type_exception" value="false"/>
<parameter key="except_block_type" value="value_matrix_row_start"/>
<parameter key="invert_selection" value="true"/>
<parameter key="include_special_attributes" value="true"/>
</operator>
<operator activated="true" class="set_role" compatibility="9.2.001" expanded="true" height="82" name="Set Role (2)" width="90" x="581" y="187">
<parameter key="attribute_name" value="Binomial_Labels_Generated"/>
<parameter key="target_role" value="label"/>
<list key="set_additional_roles"/>
</operator>
<operator activated="true" class="concurrency:cross_validation" compatibility="9.2.001" expanded="true" height="145" name="Cross Validation (2)" width="90" x="715" y="187">
<parameter key="split_on_batch_attribute" value="false"/>
<parameter key="leave_one_out" value="false"/>
<parameter key="number_of_folds" value="10"/>
<parameter key="sampling_type" value="automatic"/>
<parameter key="use_local_random_seed" value="false"/>
<parameter key="local_random_seed" value="1992"/>
<parameter key="enable_parallel_execution" value="true"/>
<process expanded="true">
<operator activated="true" class="concurrency:parallel_decision_tree" compatibility="9.2.001" expanded="true" height="103" name="Decision Tree (2)" width="90" x="112" y="34">
<parameter key="criterion" value="gain_ratio"/>
<parameter key="maximal_depth" value="10"/>
<parameter key="apply_pruning" value="true"/>
<parameter key="confidence" value="0.1"/>
<parameter key="apply_prepruning" value="true"/>
<parameter key="minimal_gain" value="0.01"/>
<parameter key="minimal_leaf_size" value="2"/>
<parameter key="minimal_size_for_split" value="4"/>
<parameter key="number_of_prepruning_alternatives" value="3"/>
</operator>
<connect from_port="training set" to_op="Decision Tree (2)" to_port="training set"/>
<connect from_op="Decision Tree (2)" from_port="model" to_port="model"/>
<portSpacing port="source_training set" spacing="0"/>
<portSpacing port="sink_model" spacing="0"/>
<portSpacing port="sink_through 1" spacing="0"/>
</process>
<process expanded="true">
<operator activated="true" class="apply_model" compatibility="9.2.001" expanded="true" height="82" name="Apply Model (2)" width="90" x="112" y="34">
<list key="application_parameters"/>
<parameter key="create_view" value="false"/>
</operator>
<operator activated="true" class="performance_binominal_classification" compatibility="9.2.001" expanded="true" height="82" name="Performance (2)" width="90" x="246" y="34">
<parameter key="main_criterion" value="first"/>
<parameter key="accuracy" value="true"/>
<parameter key="classification_error" value="false"/>
<parameter key="kappa" value="false"/>
<parameter key="AUC (optimistic)" value="false"/>
<parameter key="AUC" value="true"/>
<parameter key="AUC (pessimistic)" value="false"/>
<parameter key="precision" value="false"/>
<parameter key="recall" value="false"/>
<parameter key="lift" value="false"/>
<parameter key="fallout" value="false"/>
<parameter key="f_measure" value="false"/>
<parameter key="false_positive" value="false"/>
<parameter key="false_negative" value="false"/>
<parameter key="true_positive" value="false"/>
<parameter key="true_negative" value="false"/>
<parameter key="sensitivity" value="false"/>
<parameter key="specificity" value="false"/>
<parameter key="youden" value="false"/>
<parameter key="positive_predictive_value" value="false"/>
<parameter key="negative_predictive_value" value="false"/>
<parameter key="psep" value="false"/>
<parameter key="skip_undefined_labels" value="true"/>
<parameter key="use_example_weights" value="true"/>
</operator>
<connect from_port="model" to_op="Apply Model (2)" to_port="model"/>
<connect from_port="test set" to_op="Apply Model (2)" to_port="unlabelled data"/>
<connect from_op="Apply Model (2)" from_port="labelled data" to_op="Performance (2)" to_port="labelled data"/>
<connect from_op="Performance (2)" from_port="performance" to_port="performance 1"/>
<portSpacing port="source_model" spacing="0"/>
<portSpacing port="source_test set" spacing="0"/>
<portSpacing port="source_through 1" spacing="0"/>
<portSpacing port="sink_test set results" spacing="0"/>
<portSpacing port="sink_performance 1" spacing="0"/>
<portSpacing port="sink_performance 2" spacing="0"/>
</process>
</operator>
<operator activated="true" class="generate_attributes" compatibility="9.2.001" expanded="true" height="82" name="Generate Attributes (3)" width="90" x="313" y="289">
<list key="function_descriptions">
<parameter key="Binomial_Labels_Generated" value="if(label == "Iris-setosa", "A", (if(label == "Iris-versicolor","A",label)))"/>
</list>
<parameter key="keep_all" value="true"/>
</operator>
<operator activated="true" class="select_attributes" compatibility="9.2.001" expanded="true" height="82" name="Select Attributes (3)" width="90" x="447" y="340">
<parameter key="attribute_filter_type" value="single"/>
<parameter key="attribute" value="label"/>
<parameter key="attributes" value=""/>
<parameter key="use_except_expression" value="false"/>
<parameter key="value_type" value="attribute_value"/>
<parameter key="use_value_type_exception" value="false"/>
<parameter key="except_value_type" value="time"/>
<parameter key="block_type" value="attribute_block"/>
<parameter key="use_block_type_exception" value="false"/>
<parameter key="except_block_type" value="value_matrix_row_start"/>
<parameter key="invert_selection" value="true"/>
<parameter key="include_special_attributes" value="true"/>
</operator>
<operator activated="true" class="set_role" compatibility="9.2.001" expanded="true" height="82" name="Set Role (3)" width="90" x="581" y="340">
<parameter key="attribute_name" value="Binomial_Labels_Generated"/>
<parameter key="target_role" value="label"/>
<list key="set_additional_roles"/>
</operator>
<operator activated="true" class="concurrency:cross_validation" compatibility="9.2.001" expanded="true" height="145" name="Cross Validation (3)" width="90" x="715" y="340">
<parameter key="split_on_batch_attribute" value="false"/>
<parameter key="leave_one_out" value="false"/>
<parameter key="number_of_folds" value="10"/>
<parameter key="sampling_type" value="automatic"/>
<parameter key="use_local_random_seed" value="false"/>
<parameter key="local_random_seed" value="1992"/>
<parameter key="enable_parallel_execution" value="true"/>
<process expanded="true">
<operator activated="true" class="concurrency:parallel_decision_tree" compatibility="9.2.001" expanded="true" height="103" name="Decision Tree (3)" width="90" x="112" y="34">
<parameter key="criterion" value="gain_ratio"/>
<parameter key="maximal_depth" value="10"/>
<parameter key="apply_pruning" value="true"/>
<parameter key="confidence" value="0.1"/>
<parameter key="apply_prepruning" value="true"/>
<parameter key="minimal_gain" value="0.01"/>
<parameter key="minimal_leaf_size" value="2"/>
<parameter key="minimal_size_for_split" value="4"/>
<parameter key="number_of_prepruning_alternatives" value="3"/>
</operator>
<connect from_port="training set" to_op="Decision Tree (3)" to_port="training set"/>
<connect from_op="Decision Tree (3)" from_port="model" to_port="model"/>
<portSpacing port="source_training set" spacing="0"/>
<portSpacing port="sink_model" spacing="0"/>
<portSpacing port="sink_through 1" spacing="0"/>
</process>
<process expanded="true">
<operator activated="true" class="apply_model" compatibility="9.2.001" expanded="true" height="82" name="Apply Model (3)" width="90" x="112" y="34">
<list key="application_parameters"/>
<parameter key="create_view" value="false"/>
</operator>
<operator activated="true" class="performance_binominal_classification" compatibility="9.2.001" expanded="true" height="82" name="Performance (3)" width="90" x="246" y="34">
<parameter key="main_criterion" value="first"/>
<parameter key="accuracy" value="true"/>
<parameter key="classification_error" value="false"/>
<parameter key="kappa" value="false"/>
<parameter key="AUC (optimistic)" value="false"/>
<parameter key="AUC" value="true"/>
<parameter key="AUC (pessimistic)" value="false"/>
<parameter key="precision" value="false"/>
<parameter key="recall" value="false"/>
<parameter key="lift" value="false"/>
<parameter key="fallout" value="false"/>
<parameter key="f_measure" value="false"/>
<parameter key="false_positive" value="false"/>
<parameter key="false_negative" value="false"/>
<parameter key="true_positive" value="false"/>
<parameter key="true_negative" value="false"/>
<parameter key="sensitivity" value="false"/>
<parameter key="specificity" value="false"/>
<parameter key="youden" value="false"/>
<parameter key="positive_predictive_value" value="false"/>
<parameter key="negative_predictive_value" value="false"/>
<parameter key="psep" value="false"/>
<parameter key="skip_undefined_labels" value="true"/>
<parameter key="use_example_weights" value="true"/>
</operator>
<connect from_port="model" to_op="Apply Model (3)" to_port="model"/>
<connect from_port="test set" to_op="Apply Model (3)" to_port="unlabelled data"/>
<connect from_op="Apply Model (3)" from_port="labelled data" to_op="Performance (3)" to_port="labelled data"/>
<connect from_op="Performance (3)" from_port="performance" to_port="performance 1"/>
<portSpacing port="source_model" spacing="0"/>
<portSpacing port="source_test set" spacing="0"/>
<portSpacing port="source_through 1" spacing="0"/>
<portSpacing port="sink_test set results" spacing="0"/>
<portSpacing port="sink_performance 1" spacing="0"/>
<portSpacing port="sink_performance 2" spacing="0"/>
</process>
</operator>
<operator activated="true" class="converters:roc_curve_2_example_set" compatibility="0.5.000" expanded="true" height="82" name="ROC Curve to ExampleSet (3)" width="90" x="849" y="340">
<parameter key="Number of Points" value="500"/>
</operator>
<operator activated="true" class="converters:roc_curve_2_example_set" compatibility="0.5.000" expanded="true" height="82" name="ROC Curve to ExampleSet (2)" width="90" x="849" y="238">
<parameter key="Number of Points" value="500"/>
</operator>
<operator activated="true" class="concurrency:join" compatibility="9.2.001" expanded="true" height="82" name="Join" width="90" x="916" y="136">
<parameter key="remove_double_attributes" value="false"/>
<parameter key="join_type" value="outer"/>
<parameter key="use_id_attribute_as_key" value="false"/>
<list key="key_attributes">
<parameter key="false positive rate" value="false positive rate"/>
</list>
<parameter key="keep_both_join_attributes" value="false"/>
</operator>
<operator activated="true" class="concurrency:join" compatibility="9.2.001" expanded="true" height="82" name="Join (2)" width="90" x="1050" y="187">
<parameter key="remove_double_attributes" value="false"/>
<parameter key="join_type" value="outer"/>
<parameter key="use_id_attribute_as_key" value="false"/>
<list key="key_attributes">
<parameter key="false positive rate" value="false positive rate"/>
</list>
<parameter key="keep_both_join_attributes" value="false"/>
</operator>
<operator activated="true" class="rename" compatibility="9.2.001" expanded="true" height="82" name="Rename" width="90" x="1251" y="136">
<parameter key="old_name" value="true positive rate (Mean)"/>
<parameter key="new_name" value="true Positive Iris-versicolor"/>
<list key="rename_additional_attributes">
<parameter key="true positive rate (Mean)_from_ES2" value="true positive Iris-Setosa"/>
<parameter key="true positive rate (Mean)_from_ES2_from_ES2" value="true Positive Iris-virginica"/>
</list>
</operator>
<connect from_op="Retrieve Iris" from_port="output" to_op="Multiply" to_port="input"/>
<connect from_op="Multiply" from_port="output 1" to_op="Generate Attributes" to_port="example set input"/>
<connect from_op="Multiply" from_port="output 2" to_op="Generate Attributes (2)" to_port="example set input"/>
<connect from_op="Multiply" from_port="output 3" to_op="Generate Attributes (3)" to_port="example set input"/>
<connect from_op="Generate Attributes (2)" from_port="example set output" to_op="Select Attributes (2)" to_port="example set input"/>
<connect from_op="Generate Attributes" from_port="example set output" to_op="Select Attributes" to_port="example set input"/>
<connect from_op="Select Attributes" from_port="example set output" to_op="Set Role" to_port="example set input"/>
<connect from_op="Set Role" from_port="example set output" to_op="Cross Validation" to_port="example set"/>
<connect from_op="Cross Validation" from_port="performance 1" to_op="ROC Curve to ExampleSet" to_port="performance"/>
<connect from_op="ROC Curve to ExampleSet" from_port="example set" to_op="Join" to_port="left"/>
<connect from_op="ROC Curve to ExampleSet" from_port="original" to_port="result 3"/>
<connect from_op="Select Attributes (2)" from_port="example set output" to_op="Set Role (2)" to_port="example set input"/>
<connect from_op="Set Role (2)" from_port="example set output" to_op="Cross Validation (2)" to_port="example set"/>
<connect from_op="Cross Validation (2)" from_port="performance 1" to_op="ROC Curve to ExampleSet (2)" to_port="performance"/>
<connect from_op="Generate Attributes (3)" from_port="example set output" to_op="Select Attributes (3)" to_port="example set input"/>
<connect from_op="Select Attributes (3)" from_port="example set output" to_op="Set Role (3)" to_port="example set input"/>
<connect from_op="Set Role (3)" from_port="example set output" to_op="Cross Validation (3)" to_port="example set"/>
<connect from_op="Cross Validation (3)" from_port="performance 1" to_op="ROC Curve to ExampleSet (3)" to_port="performance"/>
<connect from_op="ROC Curve to ExampleSet (3)" from_port="example set" to_op="Join (2)" to_port="right"/>
<connect from_op="ROC Curve to ExampleSet (3)" from_port="original" to_port="result 4"/>
<connect from_op="ROC Curve to ExampleSet (2)" from_port="example set" to_op="Join" to_port="right"/>
<connect from_op="ROC Curve to ExampleSet (2)" from_port="original" to_port="result 1"/>
<connect from_op="Join" from_port="join" to_op="Join (2)" to_port="left"/>
<connect from_op="Join (2)" from_port="join" to_op="Rename" to_port="example set input"/>
<connect from_op="Rename" from_port="example set output" to_port="result 2"/>
<portSpacing port="source_input 1" spacing="0"/>
<portSpacing port="sink_result 1" spacing="0"/>
<portSpacing port="sink_result 2" spacing="0"/>
<portSpacing port="sink_result 3" spacing="0"/>
<portSpacing port="sink_result 4" spacing="0"/>
<portSpacing port="sink_result 5" spacing="0"/>
</process>
</operator>
</process>Here is the image for reference. I used multiple operators like Joins and Rename to get plots on the same figure. Once you run the process, you need to go to Example(Rename) and then select visualizations, here you need to get all the true positive rates (mean) and plot it using a line plot.
Here are the ROC curves in same plot.
Hope this helpsRegards,
Varun
https://www.varunmandalapu.com/Be Safe. Follow precautions and Maintain Social Distancing
7


Answers
Scott
Just a thought, Can't we relabel one vs all manually (converting to binomial) and then run the validation methods on different one vs all to get ROC curves and average the performance metrics? I am not sure if there is an easier way in RM.
Thanks,
Varun
Varun
https://www.varunmandalapu.com/
Be Safe. Follow precautions and Maintain Social Distancing
Lindon Ventures
Data Science Consulting from Certified RapidMiner Experts
Varun
https://www.varunmandalapu.com/
Be Safe. Follow precautions and Maintain Social Distancing
Varun
https://www.varunmandalapu.com/
Be Safe. Follow precautions and Maintain Social Distancing